| Lipoprotein lipase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.1.1.34 | ||||||||
| CAS no. | 9004-02-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
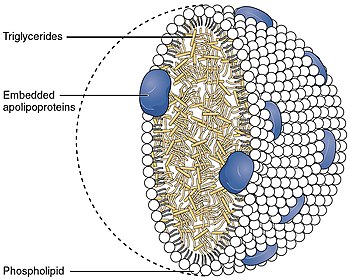
Lipoprotein lipase (LPL) (EC 3.1.1.34, systematic name triacylglycerol acylhydrolase (lipoprotein-dependent)) is a member of the lipase gene family, which includes pancreatic lipase, hepatic lipase, and endothelial lipase. It is a water-soluble enzyme that hydrolyzes triglycerides in lipoproteins, such as those found in chylomicrons and very low-density lipoproteins (VLDL), into two free fatty acids and one monoacylglycerol molecule:
- triacylglycerol + H2O = diacylglycerol + a carboxylate
It is also involved in promoting the cellular uptake of chylomicron remnants, cholesterol-rich lipoproteins, and free fatty acids.[5][6][7] LPL requires ApoC-II as a cofactor.[8][9]
LPL is attached to the luminal surface of endothelial cells in capillaries by the protein glycosylphosphatidylinositol HDL-binding protein 1 (GPIHBP1) and by heparan sulfated peptidoglycans.[10] It is most widely distributed in adipose, heart, and skeletal muscle tissue, as well as in lactating mammary glands.[11][12][13]
Synthesis
[edit]In brief, LPL is secreted from heart, muscle and adipose parenchymal cells as a glycosylated homodimer, after which it is translocated through the extracellular matrix and across endothelial cells to the capillary lumen. After translation, the newly synthesized protein is glycosylated in the endoplasmic reticulum. The glycosylation sites of LPL are Asn-43, Asn-257, and Asn-359.[5] Glucosidases then remove terminal glucose residues; it was once believed that this glucose trimming is responsible for the conformational change needed for LPL to form homodimers and become catalytically active.[5][13][14][15] In the Golgi apparatus, the oligosaccharides are further altered to result in either two complex chains, or two complex and one high-mannose chain.[5][13] In the final protein, carbohydrates account for about 12% of the molecular mass (55-58 kDa).[5][13][16]
Homodimerization is required before LPL can be secreted from cells.[16][17] After secretion, LPL is carried across endothelial cells and presented into the capillary lumen by the protein glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1.[18][19]
Structure
[edit]Crystal structures of LPL complexed with GPIHBP1 have been reported.[20][21] LPL is composed of two distinct regions: the larger N-terminus domain that contains the lipolytic active site, and the smaller C-terminus domain. These two regions are attached by a peptide linker. The N-terminus domain has an α/β hydrolase fold, which is a globular structure containing a central β sheet surrounded by α helices. The C-terminus domain is a β sandwich formed by two β sheet layers, and resembles an elongated cylinder.
Mechanism
[edit]
The active site of LPL is composed of the conserved Ser-132, Asp-156, and His-241 triad. Other important regions of the N-terminal domain for catalysis includes an oxyanion hole (Trp-55, Leu-133), a lid region (residues 216-239), as well as a β5 loop (residues 54-64).[5][11][15] The ApoC-II binding site is currently unknown, but it is predicted that residues on both N-and C-terminal domains are necessary for this interaction to occur. The C-terminal domain appears to confer LPL’s substrate specificity; it has a higher affinity for large triacylglyceride-rich lipoproteins than cholesterol-rich lipoproteins.[22] The C-terminal domain is also important for binding to LDL’s receptors.[23] Both the N-and C-terminal domains contain heparin binding sites distal to the lipid binding sites; LPL therefore serves as a bridge between the cell surface and lipoproteins. Importantly, LPL binding to the cell surface or receptors is not dependent on its catalytic activity.[24]
The LPL non-covalent homodimer has a head-to-tail arrangement of the monomers. The Ser/Asp/His triad is in a hydrophobic groove that is blocked from solvent by the lid.[5][11] Upon binding to ApoC-II and lipid in the lipoprotein, the C-terminal domain presents the lipid substrate to the lid region. The lipid interacts with both the lid region and the hydrophobic groove at the active site; this causes the lid to move, providing access to the active site. The β5 loop folds back into the protein core, bringing one of the electrophiles of the oxyanion hole into position for lipolysis.[5] The glycerol backbone of the lipid is then able to enter the active site and is hydrolyzed.
Two molecules of ApoC-II can attach to each LPL dimer.[25] It is estimated that up to forty LPL dimers may act simultaneously on a single lipoprotein.[5] In regard to kinetics, it is believed that release of product into circulation is the rate-limiting step in the reaction.[11]
Function
[edit]LPL gene encodes lipoprotein lipase, which is expressed in the heart, muscle, and adipose tissue.[26][27] LPL functions as a homodimer, and has the dual functions of triglyceride hydrolase and ligand/bridging factor for receptor-mediated lipoprotein uptake. Through catalysis, VLDL is converted to IDL and then to LDL. Severe mutations that cause LPL deficiency result in type I hyperlipoproteinemia, while less extreme mutations in LPL are linked to many disorders of lipoprotein metabolism.[28]
Regulation
[edit]LPL is controlled transcriptionally and posttranscriptionally.[29] The circadian clock may be important in the control of Lpl mRNA levels in peripheral tissues.[30]
LPL isozymes are regulated differently depending on the tissue. For example, insulin is known to activate LPL in adipocytes and its placement in the capillary endothelium. By contrast, insulin has been shown to decrease expression of muscle LPL.[31] Muscle and myocardial LPL is instead activated by glucagon and adrenaline. This helps to explain why during fasting, LPL activity increases in muscle tissue and decreases in adipose tissue, whereas after a meal, the opposite occurs.[5][13]
Consistent with this, dietary macronutrients differentially affect adipose and muscle LPL activity. After 16 days on a high-carbohydrate or a high-fat diet, LPL activity increased significantly in both tissues 6 hours after a meal of either composition, but there was a significantly greater rise in adipose tissue LPL in response to the high-carbohydrate diet compared to the high-fat diet. There was no difference between the two diets' effects on insulin sensitivity or fasting LPL activity in either tissue.[32]
The concentration of LPL displayed on endothelial cell surface cannot be regulated by endothelial cells, as they neither synthesize nor degrade LPL. Instead, this regulation occurs by managing the flux of LPL arriving at the lipolytic site and by regulating the activity of LPL present on the endothelium. A key protein involved in controlling the activity of LPL is ANGPTL4, which serves as a local inhibitor of LPL. Induction of ANGPTL4 accounts for the inhibition of LPL activity in white adipose tissue during fasting. Growing evidence implicates ANGPTL4 in the physiological regulation of LPL activity in a variety of tissues.[33]
An ANGPTL3-4-8 model was proposed to explain the variations of LPL activity during the fed-fast cycle.[34] Specifically, feeding induces ANGPTL8, activating the ANGPTL8–ANGPTL3 pathway, which inhibits LPL in cardiac and skeletal muscles, thereby making circulating triglycerides available for uptake by white adipose tissue, in which LPL activity is elevated owing to diminished ANGPTL4; the reverse is true during fasting, which suppresses ANGPTL8 but induces ANGPTL4, thereby directing triglycerides to muscles. The model suggests a general framework for how triglyceride trafficking is regulated.[34]
Clinical significance
[edit]Lipoprotein lipase deficiency leads to hypertriglyceridemia (elevated levels of triglycerides in the bloodstream).[35] In mice, overexpression of LPL has been shown to cause insulin resistance,[36][37] and to promote obesity.[30]
A high adipose tissue LPL response to a high-carbohydrate diet may predispose toward fat gain. One study reported that subjects gained more body fat over the next four years if, after following a high-carbohydrate diet and partaking of a high-carbohydrate meal, they responded with an increase in adipose tissue LPL activity per adipocyte, or a decrease in skeletal muscle LPL activity per gram of tissue.[38]
LPL expression has been shown to be a prognostic predictor in Chronic lymphocytic leukemia.[39] In this haematological disorder, LPL appears to provide fatty acids as an energy source to malignant cells.[40] Thus, elevated levels of LPL mRNA or protein are considered to be indicators of poor prognosis.[41][42][43][44][45][46][47][48][49][50]
Interactions
[edit]Lipoprotein lipase has been shown to interact with LRP1.[51][52][53] It is also a ligand for α2M, GP330, and VLDL receptors.[23] LPL has been shown to be a ligand for LRP2, albeit at a lower affinity than for other receptors; however, most of the LPL-dependent VLDL degradation can be attributed to the LRP2 pathway.[23] In each case, LPL serves as a bridge between receptor and lipoprotein. While LPL is activated by ApoC-II, it is inhibited by ApoCIII.[11]
In other organisms
[edit]The LPL gene is highly conserved across vertebrates. Lipoprotein lipase is involved in lipid transport in the placentae of live bearing lizards (Pseudemoia entrecasteauxii).[54]
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000175445 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000015568 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c d e f g h i j Mead JR, Irvine SA, Ramji DP (December 2002). "Lipoprotein lipase: structure, function, regulation, and role in disease". J. Mol. Med. 80 (12): 753–69. doi:10.1007/s00109-002-0384-9. PMID 12483461. S2CID 40089672.
- ^ Rinninger F, Kaiser T, Mann WA, Meyer N, Greten H, Beisiegel U (July 1998). "Lipoprotein lipase mediates an increase in the selective uptake of high density lipoprotein-associated cholesteryl esters by hepatic cells in culture". J. Lipid Res. 39 (7): 1335–48. doi:10.1016/S0022-2275(20)32514-1. PMID 9684736.
- ^ Ma Y, Henderson HE, Liu MS, Zhang H, Forsythe IJ, Clarke-Lewis I, Hayden MR, Brunzell JD (November 1994). "Mutagenesis in four candidate heparin binding regions (residues 279-282, 291-304, 390-393, and 439-448) and identification of residues affecting heparin binding of human lipoprotein lipase". J. Lipid Res. 35 (11): 2049–59. doi:10.1016/S0022-2275(20)39951-X. PMID 7868983.
- ^ Kim SY, Park SM, Lee ST (January 2006). "Apolipoprotein C-II is a novel substrate for matrix metalloproteinases". Biochem. Biophys. Res. Commun. 339 (1): 47–54. doi:10.1016/j.bbrc.2005.10.182. PMID 16314153.
- ^ Kinnunen PK, Jackson RL, Smith LC, Gotto AM, Sparrow JT (November 1977). "Activation of lipoprotein lipase by native and synthetic fragments of human plasma apolipoprotein C-II". Proc. Natl. Acad. Sci. U.S.A. 74 (11): 4848–51. Bibcode:1977PNAS...74.4848K. doi:10.1073/pnas.74.11.4848. PMC 432053. PMID 270715.
- ^ Meneghetti MC, Hughes AJ, Rudd TR, Nader HB, Powell AK, Yates EA, Lima MA (September 2015). "Heparan sulfate and heparin interactions with proteins". Journal of the Royal Society, Interface. 12 (110): 0589. doi:10.1098/rsif.2015.0589. PMC 4614469. PMID 26289657.
- ^ a b c d e Wang CS, Hartsuck J, McConathy WJ (January 1992). "Structure and functional properties of lipoprotein lipase" (PDF). Biochimica et Biophysica Acta. 1123 (1): 1–17. doi:10.1016/0005-2728(92)90119-M. PMID 1730040.
- ^ Wong H, Schotz MC (July 2002). "The lipase gene family". Journal of Lipid Research. 43 (7): 993–9. doi:10.1194/jlr.R200007-JLR200. PMID 12091482.
- ^ a b c d e Braun JE, Severson DL (October 1992). "Regulation of the synthesis, processing and translocation of lipoprotein lipase". The Biochemical Journal. 287 ( Pt 2) (2): 337–47. doi:10.1042/bj2870337. PMC 1133170. PMID 1445192.
- ^ Semb H, Olivecrona T (March 1989). "The relation between glycosylation and activity of guinea pig lipoprotein lipase". J. Biol. Chem. 264 (7): 4195–200. doi:10.1016/S0021-9258(19)84982-7. PMID 2521859.
- ^ a b Wong H, Davis RC, Thuren T, Goers JW, Nikazy J, Waite M, Schotz MC (April 1994). "Lipoprotein lipase domain function". J. Biol. Chem. 269 (14): 10319–23. doi:10.1016/S0021-9258(17)34063-2. PMID 8144612.
- ^ a b Vannier C, Ailhaud G (August 1989). "Biosynthesis of lipoprotein lipase in cultured mouse adipocytes. II. Processing, subunit assembly, and intracellular transport". J. Biol. Chem. 264 (22): 13206–16. doi:10.1016/S0021-9258(18)51616-1. PMID 2753912.
- ^ Ong JM, Kern PA (February 1989). "The role of glucose and glycosylation in the regulation of lipoprotein lipase synthesis and secretion in rat adipocytes". J. Biol. Chem. 264 (6): 3177–82. doi:10.1016/S0021-9258(18)94047-0. PMID 2644281.
- ^ Beigneux AP, Davies BS, Gin P, Weinstein MM, Farber E, Qiao X, Peale F, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, de Sauvage F, Ryan RO, Fong LG, Bensadoun A, Young SG (2007). "Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons". Cell Metabolism. 5 (4): 279–291. doi:10.1016/j.cmet.2007.02.002. PMC 1913910. PMID 17403372.
- ^ Davies BS, Beigneux AP, Barnes RH, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyrén R, Goldberg I, Olivecrona G, Bensadoun A, Young SG, Fong LG (July 2010). "GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries". Cell Metabolism. 12 (1): 42–52. doi:10.1016/j.cmet.2010.04.016. PMC 2913606. PMID 20620994.
- ^ PDB: 6E7K; Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, et al. (January 2019). "Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis". Proceedings of the National Academy of Sciences of the United States of America. 116 (5): 1723–1732. Bibcode:2019PNAS..116.1723B. doi:10.1073/pnas.1817984116. PMC 6358717. PMID 30559189.
- ^ PDB: 6OAU, 6OAZ, 6OB0; Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, et al. (May 2019). "Structure of lipoprotein lipase in complex with GPIHBP1". Proceedings of the National Academy of Sciences of the United States of America. 116 (21): 10360–10365. Bibcode:2019PNAS..11610360A. doi:10.1073/pnas.1820171116. PMC 6534989. PMID 31072929.
- ^ Lookene A, Nielsen MS, Gliemann J, Olivecrona G (April 2000). "Contribution of the carboxy-terminal domain of lipoprotein lipase to interaction with heparin and lipoproteins". Biochem. Biophys. Res. Commun. 271 (1): 15–21. doi:10.1006/bbrc.2000.2530. PMID 10777674.
- ^ a b c Medh JD, Bowen SL, Fry GL, Ruben S, Andracki M, Inoue I, Lalouel JM, Strickland DK, Chappell DA (July 1996). "Lipoprotein lipase binds to low density lipoprotein receptors and induces receptor-mediated catabolism of very low density lipoproteins in vitro". J. Biol. Chem. 271 (29): 17073–80. doi:10.1074/jbc.271.29.17073. PMID 8663292.
- ^ Beisiegel U, Weber W, Bengtsson-Olivecrona G (October 1991). "Lipoprotein lipase enhances the binding of chylomicrons to low density lipoprotein receptor-related protein". Proc. Natl. Acad. Sci. U.S.A. 88 (19): 8342–6. Bibcode:1991PNAS...88.8342B. doi:10.1073/pnas.88.19.8342. PMC 52504. PMID 1656440.
- ^ McIlhargey TL, Yang Y, Wong H, Hill JS (June 2003). "Identification of a lipoprotein lipase cofactor-binding site by chemical cross-linking and transfer of apolipoprotein C-II-responsive lipolysis from lipoprotein lipase to hepatic lipase". J. Biol. Chem. 278 (25): 23027–35. doi:10.1074/jbc.M300315200. PMID 12682050.
- ^ Protein Atlas, Protein Atlas. "Tissue expression of LPL - Summary - The Human Protein Atlas". www.proteinatlas.org. The Human Protein Atlas. Retrieved 25 July 2019.
- ^ Gene Cards, Gene Cards. "Human Gene Database". www.genecards.org. GeneCardsSuite. Retrieved 25 July 2019.
- ^ "Entrez Gene: LPL lipoprotein lipase".
- ^ Wang H, Eckel RH (2009). "Lipoprotein lipase: from gene to obesity". Am J Physiol Endocrinol Metab. 297 (2): E271–88. doi:10.1152/ajpendo.90920.2008. PMID 19318514.
- ^ a b Delezie J, Dumont S, Dardente H, Oudart H, Gréchez-Cassiau A, Klosen P, et al. (2012). "The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism". FASEB J. 26 (8): 3321–35. doi:10.1096/fj.12-208751. PMID 22562834. S2CID 31204290.
- ^ Kiens B, Lithell H, Mikines KJ, Richter EA (October 1989). "Effects of insulin and exercise on muscle lipoprotein lipase activity in man and its relation to insulin action". J. Clin. Invest. 84 (4): 1124–9. doi:10.1172/JCI114275. PMC 329768. PMID 2677048.
- ^ Yost TJ, Jensen DR, Haugen BR, Eckel RH (August 1998). "Effect of dietary macronutrient composition on tissue-specific lipoprotein lipase activity and insulin action in normal-weight subjects" (PDF). Am. J. Clin. Nutr. 68 (2): 296–302. doi:10.1093/ajcn/68.2.296. PMID 9701186.
- ^ Dijk W, Kersten S (2014). "Regulation of lipoprotein lipase by Angptl4". Trends Endocrinol. Metab. 25 (3): 146–155. doi:10.1016/j.tem.2013.12.005. PMID 24397894. S2CID 10273285.
- ^ a b Zhang R (April 2016). "The ANGPTL3-4-8 model, a molecular mechanism for triglyceride trafficking". Open Biol. 6 (4): 150272. doi:10.1098/rsob.150272. PMC 4852456. PMID 27053679.
- ^ Okubo M, Horinishi A, Saito M, Ebara T, Endo Y, Kaku K, Murase T, Eto M (November 2007). "A novel complex deletion-insertion mutation mediated by Alu repetitive elements leads to lipoprotein lipase deficiency". Mol. Genet. Metab. 92 (3): 229–33. doi:10.1016/j.ymgme.2007.06.018. PMID 17706445.
- ^ Ferreira LD, Pulawa LK, Jensen DR, Eckel RH (2001). "Overexpressing human lipoprotein lipase in mouse skeletal muscle is associated with insulin resistance". Diabetes. 50 (5): 1064–8. doi:10.2337/diabetes.50.5.1064. PMID 11334409.
- ^ Kim JK, Fillmore JJ, Chen Y, Yu C, Moore IK, Pypaert M, et al. (2001). "Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance". Proc Natl Acad Sci U S A. 98 (13): 7522–7. Bibcode:2001PNAS...98.7522K. doi:10.1073/pnas.121164498. PMC 34701. PMID 11390966.
- ^ Ferland A, Château-Degat ML, Hernandez TL, Eckel RH (May 2012). "Tissue-specific responses of lipoprotein lipase to dietary macronutrient composition as a predictor of weight gain over 4 years". Obesity (Silver Spring). 20 (5): 1006–11. doi:10.1038/oby.2011.372. PMID 22262159. S2CID 40167321.
- ^ Prieto D, Oppezzo P (December 2017). "Lipoprotein Lipase Expression in Chronic Lymphocytic Leukemia: New Insights into Leukemic Progression". Molecules. 22 (12): 2083. doi:10.3390/molecules22122083. PMC 6149886. PMID 29206143.
- ^ Rozovski U, Hazan-Halevy I, Barzilai M, Keating MJ, Estrov Z (8 December 2015). "Metabolism pathways in chronic lymphocytic leukemia". Leukemia & Lymphoma. 57 (4): 758–65. doi:10.3109/10428194.2015.1106533. PMC 4794359. PMID 26643954.
- ^ Oppezzo P, Vasconcelos Y, Settegrana C, Jeannel D, Vuillier F, Legarff-Tavernier M, Kimura EY, Bechet S, Dumas G, Brissard M, Merle-Béral H, Yamamoto M, Dighiero G, Davi F (July 2005). "The LPL/ADAM29 expression ratio is a novel prognosis indicator in chronic lymphocytic leukemia". Blood. 106 (2): 650–7. doi:10.1182/blood-2004-08-3344. PMID 15802535.
- ^ Heintel D, Kienle D, Shehata M, Kröber A, Kroemer E, Schwarzinger I, Mitteregger D, Le T, Gleiss A, Mannhalter C, Chott A, Schwarzmeier J, Fonatsch C, Gaiger A, Döhner H, Stilgenbauer S, Jäger U (July 2005). "High expression of lipoprotein lipase in poor risk B-cell chronic lymphocytic leukemia". Leukemia. 19 (7): 1216–23. doi:10.1038/sj.leu.2403748. PMID 15858619.
- ^ van't Veer MB, Brooijmans AM, Langerak AW, Verhaaf B, Goudswaard CS, Graveland WJ, van Lom K, Valk PJ (January 2006). "The predictive value of lipoprotein lipase for survival in chronic lymphocytic leukemia". Haematologica. 91 (1): 56–63. PMID 16434371.
- ^ Nückel H, Hüttmann A, Klein-Hitpass L, Schroers R, Führer A, Sellmann L, Dührsen U, Dürig J (June 2006). "Lipoprotein lipase expression is a novel prognostic factor in B-cell chronic lymphocytic leukemia". Leukemia & Lymphoma. 47 (6): 1053–61. doi:10.1080/10428190500464161. PMID 16840197. S2CID 20532204.
- ^ Mansouri M, Sevov M, Fahlgren E, Tobin G, Jondal M, Osorio L, Roos G, Olivecrona G, Rosenquist R (March 2010). "Lipoprotein lipase is differentially expressed in prognostic subsets of chronic lymphocytic leukemia but displays invariably low catalytical activity". Leukemia Research. 34 (3): 301–6. doi:10.1016/j.leukres.2009.07.032. PMID 19709746.
- ^ Kaderi MA, Kanduri M, Buhl AM, Sevov M, Cahill N, Gunnarsson R, Jansson M, Smedby KE, Hjalgrim H, Jurlander J, Juliusson G, Mansouri L, Rosenquist R (August 2011). "LPL is the strongest prognostic factor in a comparative analysis of RNA-based markers in early chronic lymphocytic leukemia". Haematologica. 96 (8): 1153–60. doi:10.3324/haematol.2010.039396. PMC 3148909. PMID 21508119.
- ^ Porpaczy E, Tauber S, Bilban M, Kostner G, Gruber M, Eder S, Heintel D, Le T, Fleiss K, Skrabs C, Shehata M, Jäger U, Vanura K (June 2013). "Lipoprotein lipase in chronic lymphocytic leukaemia - strong biomarker with lack of functional significance". Leukemia Research. 37 (6): 631–6. doi:10.1016/j.leukres.2013.02.008. PMID 23478142.
- ^ Mátrai Z, Andrikovics H, Szilvási A, Bors A, Kozma A, Ádám E, Halm G, Karászi É, Tordai A, Masszi T (January 2017). "Lipoprotein Lipase as a Prognostic Marker in Chronic Lymphocytic Leukemia". Pathology & Oncology Research. 23 (1): 165–171. doi:10.1007/s12253-016-0132-z. PMID 27757836. S2CID 22647616.
- ^ Prieto D, Seija N, Uriepero A, Souto-Padron T, Oliver C, Irigoin V, Guillermo C, Navarrete MA, Inés Landoni A, Dighiero G, Gabus R, Giordano M, Oppezzo P (August 2018). "LPL protein in Chronic Lymphocytic Leukaemia have different origins in Mutated and Unmutated patients. Advances for a new prognostic marker in CLL". British Journal of Haematology. 182 (4): 521–525. doi:10.1111/bjh.15427. hdl:11336/95516. PMID 29953583.
- ^ Rombout A, Verhasselt B, Philippé J (November 2016). "Lipoprotein lipase in chronic lymphocytic leukemia: function and prognostic implications". European Journal of Haematology. 97 (5): 409–415. doi:10.1111/ejh.12789. PMID 27504855.
- ^ Williams SE, Inoue I, Tran H, Fry GL, Pladet MW, Iverius PH, Lalouel JM, Chappell DA, Strickland DK (March 1994). "The carboxyl-terminal domain of lipoprotein lipase binds to the low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor (LRP) and mediates binding of normal very low density lipoproteins to LRP". J. Biol. Chem. 269 (12): 8653–8. doi:10.1016/S0021-9258(17)37017-5. PMID 7510694.
- ^ Nykjaer A, Nielsen M, Lookene A, Meyer N, Røigaard H, Etzerodt M, Beisiegel U, Olivecrona G, Gliemann J (December 1994). "A carboxyl-terminal fragment of lipoprotein lipase binds to the low density lipoprotein receptor-related protein and inhibits lipase-mediated uptake of lipoprotein in cells". J. Biol. Chem. 269 (50): 31747–55. doi:10.1016/S0021-9258(18)31759-9. PMID 7989348.
- ^ Chappell DA, Fry GL, Waknitz MA, Iverius PH, Williams SE, Strickland DK (December 1992). "The low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor binds and mediates catabolism of bovine milk lipoprotein lipase". J. Biol. Chem. 267 (36): 25764–7. doi:10.1016/S0021-9258(18)35675-8. PMID 1281473.
- ^ Griffith OW, Ujvari B, Belov K, Thompson MB (November 2013). "Placental lipoprotein lipase (LPL) gene expression in a placentotrophic lizard, Pseudemoia entrecasteauxii". Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 320 (7): 465–70. doi:10.1002/jez.b.22526. PMID 23939756.
Further reading
[edit]- Zechner R (1997). "The tissue-specific expression of lipoprotein lipase: implications for energy and lipoprotein metabolism". Curr. Opin. Lipidol. 8 (2): 77–88. doi:10.1097/00041433-199704000-00005. PMID 9183545.
- Fisher RM, Humphries SE, Talmud PJ (1998). "Common variation in the lipoprotein lipase gene: effects on plasma lipids and risk of atherosclerosis". Atherosclerosis. 135 (2): 145–59. doi:10.1016/S0021-9150(97)00199-8. PMID 9430364.
- Beisiegel U (1998). "Lipoprotein metabolism". Eur. Heart J. 19 Suppl A: A20–3. doi:10.1093/eurheartj/19.Abstract_Supplement.1. hdl:20.500.11820/f1238768-8077-44c8-a7f0-72d0d4b9c819. PMID 9519338.
- Pentikäinen MO, Oksjoki R, Oörni K, Kovanen PT (2002). "Lipoprotein lipase in the arterial wall: linking LDL to the arterial extracellular matrix and much more". Arterioscler. Thromb. Vasc. Biol. 22 (2): 211–7. doi:10.1161/hq0102.101551. PMID 11834518.
- Lichtenstein L, Berbée JF, van Dijk SJ, van Dijk KW, Bensadoun A, Kema IP, Voshol PJ, Müller M, Rensen PC, Kersten S (November 2007). "Angptl4 upregulates cholesterol synthesis in liver via inhibition of LPL- and HL-dependent hepatic cholesterol uptake". Arterioscler. Thromb. Vasc. Biol. 27 (11): 2420–7. doi:10.1161/ATVBAHA.107.151894. PMID 17761937.
- Lichtenstein L, Mattijssen F, de Wit NJ, Georgiadi A, Hooiveld GJ, van der Meer R, He Y, Qi L, Köster A, Tamsma JT, Tan NS, Müller M, Kersten S (December 2010). "Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages". Cell Metab. 12 (6): 580–92. doi:10.1016/j.cmet.2010.11.002. PMC 3387545. PMID 21109191.
External links
[edit]- GeneReviews/NCBI/NIH/UW entry on Familial Lipoprotein Lipase Deficiency
- Gene therapy for lipoprotein lipase deficiency
- Lipoprotein+lipase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)